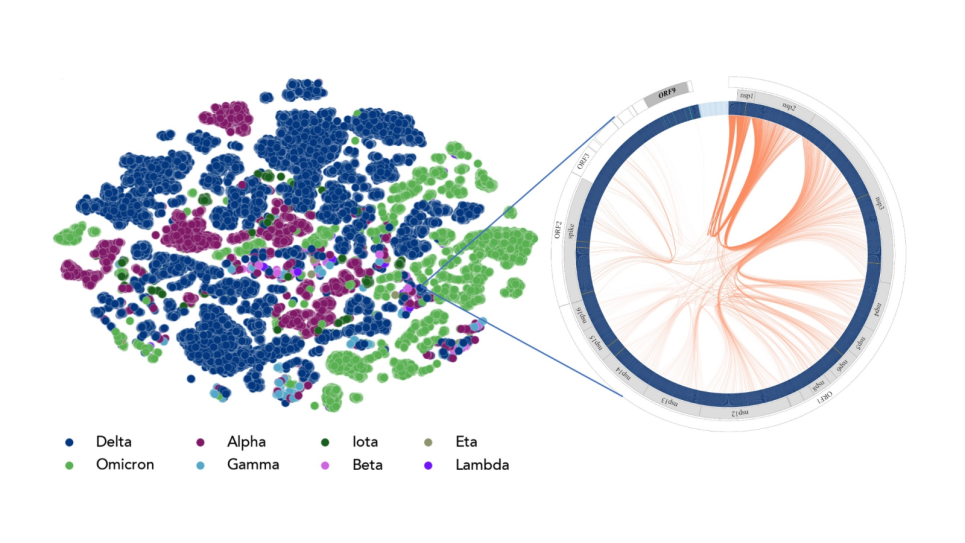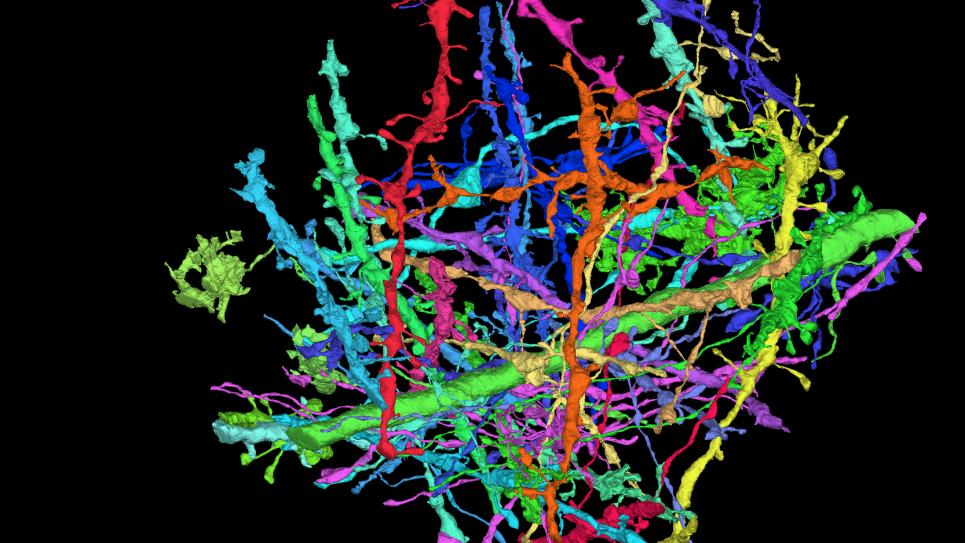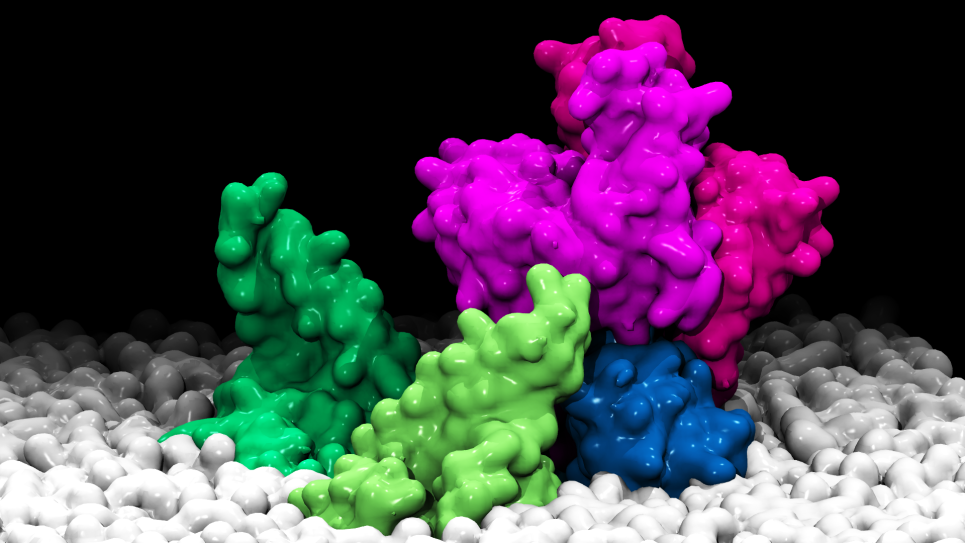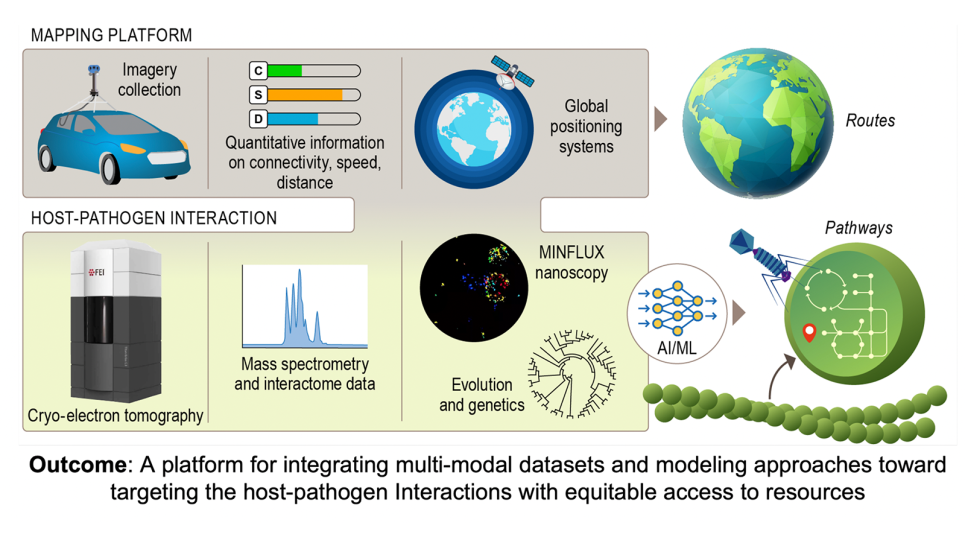
The overall concept of a “Google Earth”-like platform of a digital twin is to allow users to rapidly glance at the spatial features of host-pathogen interactions and navigate to details of interest. The platform will integrate and transform data from in situ images captured by cryo-ET, from key pathways, interactions and phenotypes mapped by multi-omics tools, and from adaptations tracked by environmental sampling and genomic analysis.