Argonne’s HPC infrastructure plays a key role in advancing the lab’s broad portfolio of research, which includes health-focused studies. Anchored by the new Aurora exascale system, the Argonne Leadership Computing Facility (ALCF) operates supercomputing and AI resources that provide the computational power to process massive datasets, train large-scale AI models, and simulate complex biological systems, paving the way for new breakthroughs in cancer research. The ALCF is a DOE Office of Science user facility that is available to researchers across the world.
Using Aurora, Brettin and colleagues led an early science project that focused on AI-driven drug discovery for cancer. Their work demonstrated how the system’s immense processing power can help accelerate the discovery of promising new drug molecules.
“The combination of AI plus Aurora allowed us to screen 50 billion small molecules for potential cancer inhibitors in about 20 minutes,” he said. “On earlier supercomputers, screening even a billion molecules in that amount of time was unthinkable.”
Taking this research further, Brettin and his team have kicked off a project supported by Advanced Research Projects Agency for Health (ARPA-H), part of the U.S. Department of Health and Human Services. Building on years of collaboration with the National Cancer Institute (NCI) and other partners, this new effort combines AI and HPC with pre-clinical laboratory testing to take their drug discovery research to the next level.
"It’s exciting to see our efforts expanding and our cancer research going strong nearly a decade since it really took off,” Brettin said. “A lot of that success comes down to Argonne’s leadership in AI, which has been key to driving this work forward."
A decade of AI-driven innovation
The origins of Argonne’s AI-driven cancer research date back to 2016, when DOE forged a partnership with NCI to employ advanced computing technologies in the fight against cancer. Argonne has been a key player in this collaboration, developing software and AI tools to accelerate progress in cancer research.
A cornerstone of this effort was the CANcer Distributed Learning Environment (CANDLE) project, a collaboration involving DOE’s Argonne, Oak Ridge, Lawrence Livermore, and Los Alamos national laboratories, and NCI’s Frederick National Laboratory for Cancer Research. Led by Argonne’s Rick Stevens and supported by DOE’s Exascale Computing Project, CANDLE’s goal was to develop a scalable deep learning software stack for the nation's exascale supercomputers.
“CANDLE was aimed at tackling the most challenging problems in cancer research,” said Stevens, Argonne associate laboratory director for Computing, Environment and Life Sciences. “We created a platform that could take full advantage of the most powerful supercomputers and all the cancer data being accumulated from lab experiments, medical records, and molecular simulations. It was designed to help spur the development of new AI models that drive cancer research forward.”
With that foundation, the lab’s focus extended from building AI models to developing a rigorous method to assess the growing number of models emerging from the broader cancer research community.
“We were blazing new ground—developing these models and putting them out there,” Brettin said. “Soon, the academic community joined in and began creating their own models to predict how tumors respond to drug treatments. That’s when NCI turned to us for help in building a framework to evaluate all the models.”
This shift led to the launch of the IMPROVE (Innovative Methodologies and New Data for Predictive Oncology Model Evaluation) project in 2021. Led by Argonne in collaboration with the Frederick National Laboratory for Cancer Research, the IMPROVE team set out to develop a standardized way to analyze and compare the performance of various drug response prediction models.
While the team’s efforts are laying the groundwork for more reliable and effective AI models, the project continues to evolve to meet new challenges as they emerge.
“We’ve recently shifted our evaluation targets from traditional AI models that predict how tumors respond to drugs to large language models designed to find new molecules that could inhibit tumor growth,” Brettin said. “We’re looking at models released in the public domain as well as some of our own.”
The IDEAL next step: “undruggable” targets
Expanding on the DOE-NCI efforts, Brettin and colleagues are now setting their sights on a longstanding challenge in cancer research: “undruggable” targets (proteins that are known to resist chemical treatments). Supported by ARPA-H, the Integrated AI and Experimental Approaches for Targeting Intrinsically Disordered Proteins in Designing Anticancer Ligands (IDEAL) project is a collaboration between Argonne and the University of Chicago Medicine Comprehensive Cancer Center.
“This project brings together stellar computational capabilities, the best structural biology resources, a world-class cancer center and some of the best scientists,” said Kunle Odunsi, MD, PhD, director of the University of Chicago Medicine Comprehensive Cancer Center; dean for Oncology, Biological Sciences Division; and The AbbVie Foundation Distinguished Service Professor of Obstetrics and Gynecology, in a press release announcing the project. “I believe this ‘dream team’ has the potential to revolutionize the cancer drug discovery timeline and change the paradigm for patients that currently have a poor prognosis and little hope for recovery.”
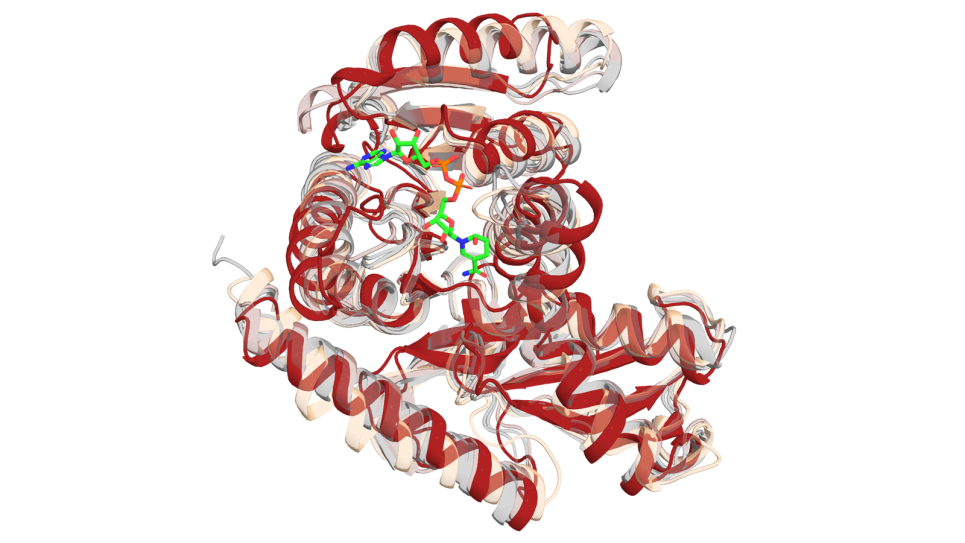
With a focus on proteins that play a key role in cancer progression, the team’s work begins with a list of targets identified through lab experiments. The researchers then retrieve the protein sequences from public databases. If a protein’s three-dimensional structure is unknown, they work with scientists at the Advanced Photon Source, a DOE Office of Science user facility, to determine it. The APS was recently upgraded to deliver significantly brighter X-ray beams, giving scientists a powerful tool to advance research across different fields.
“Using the APS’s ultrabright X-rays, we can collect data faster, capture high-resolution structural details of the proteins and visualize the binding of ligands to provide insights that help the team target them more effectively,” said Argonne Distinguished Fellow Andrzej Joachimiak.
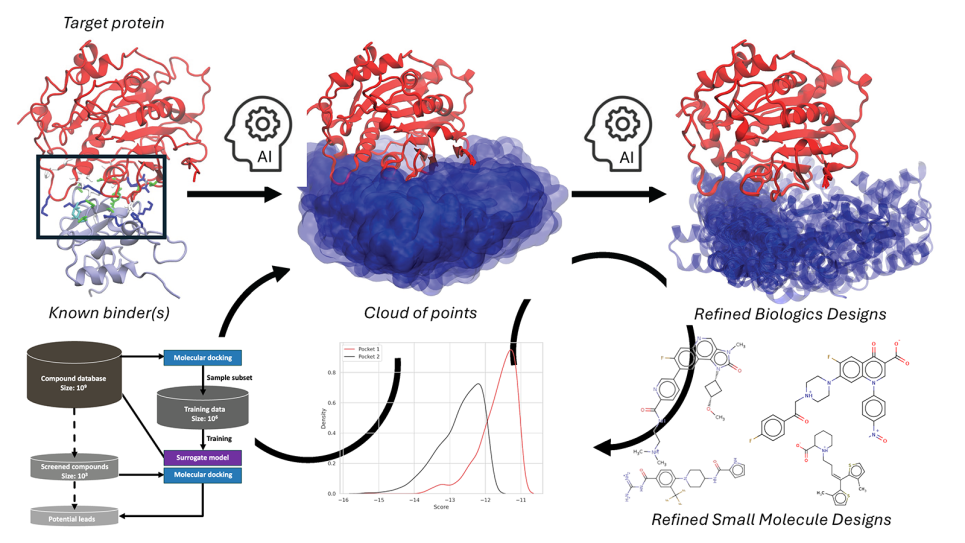
After determining the protein's structure, the team turns to Aurora to simulate the behavior and interactions of the protein at the atomic level. The simulations combined with experimental data help identify areas where small molecules might bind to inhibit the protein’s activity.
“Imagine a protein being like a piece of chewed-up gum, full of little pockets and crevices,” Brettin explained. “We simulate the structure on Aurora, find those pockets, and then search for small molecules that can bind there and stop the protein from functioning.”
The computational results are then relayed to experimental collaborators to validate the findings. “We provide a list of compounds we predict will inhibit the target protein. They order the molecules, test them in the lab, and measure their effectiveness—either by observing the protein’s activity or by monitoring tumor cell growth,” Brettin said.
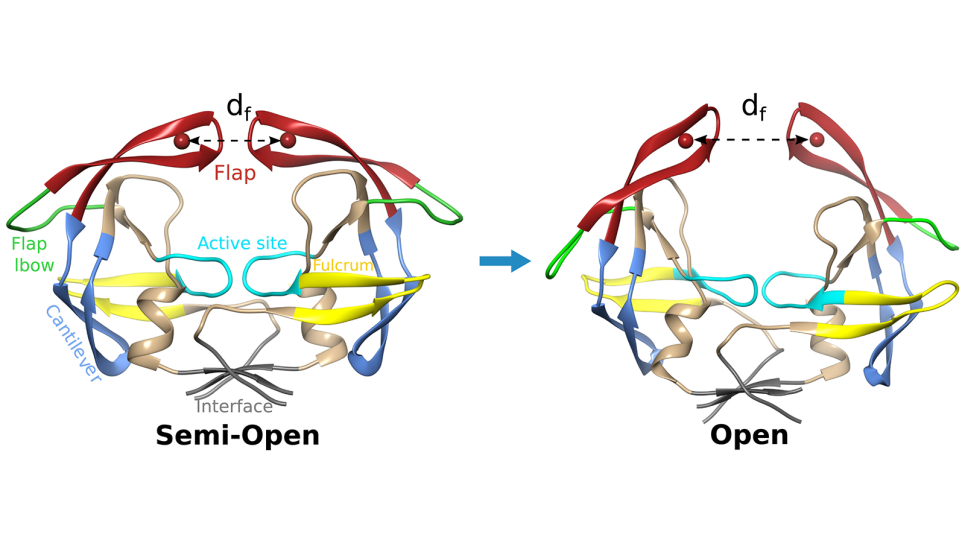
Adding to the challenge is the team’s focus on undruggable targets. Inhibiting these proteins has eluded researchers for decades, earning them a reputation as one of the most difficult problems in cancer biology.
“Historically, scientists haven’t been able to find compounds that work against these so-called undruggable proteins,” Brettin said. “But we think we can do it now because we’re bringing a couple of tools to the table that didn’t exist before, namely Aurora and AI.”